
The iterator interface demonstrated above allocates an object for each record and that may be a bottleneck of reading data from a BAM file. The size of a BAM file is often extremely large. Println(BAM.refname(record), ':', BAM.position(record)) Again, we refer you to the official for more details.Ī BAM file stores this same information but in a binary and compressible format that does not make for pretty printing here! Reading SAM and BAM filesĪ typical script iterating over all records in a file looks like below: using BioAlignments In the example above, r003 is a chimeric read, and r004 is a split alignment, and r001 are mate pair reads. Sometimes one record describes one read, but there are other cases like chimeric reads and split alignments, where multiple records apply to one read.
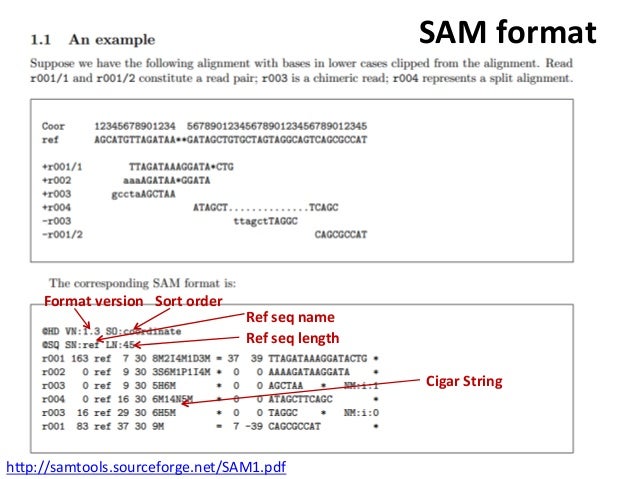
Each record describes how a read aligns to some reference sequence. Where the first two lines are part of the "header", and the following lines are "records". Ī very very simple SAM file looks like the following: VN:1.6 SN:ref LN:45 If you have questions about the SAM and BAM formats or any of the terminology used when discussing these formats, see the published, which is maintained by the. BioAlignments provides several data formats commonly used for this kind of task.īioAlignments offers high-performance tools for SAM and BAM file formats, which are the most popular file formats. One of the most common tasks in bioinformatics is to align these reads against known reference genomes, chromosomes, or contigs.

High-throughput sequencing (HTS) technologies generate a large amount of data in the form of a large number of nucleotide sequencing reads.


 0 kommentar(er)
0 kommentar(er)
